Unveiling the Hidden Variability in K562 Cell Lines: New Insights from Recent Research
The K-562 cell line, a pivotal model in leukemia research, has been a cornerstone in understanding chronic myeloid leukemia (CML) for years. However, recent findings highlight significant genetic and phenotypic variability among K562 sublines, challenging assumptions of uniformity. This blog explores new insights from the recent Scientific Reports (2024) study, showcasing the implications of these variations in scientific experiments.
Key Findings on K562 Cell Line Variability
-
Genomic Variability in K562 Subtypes:
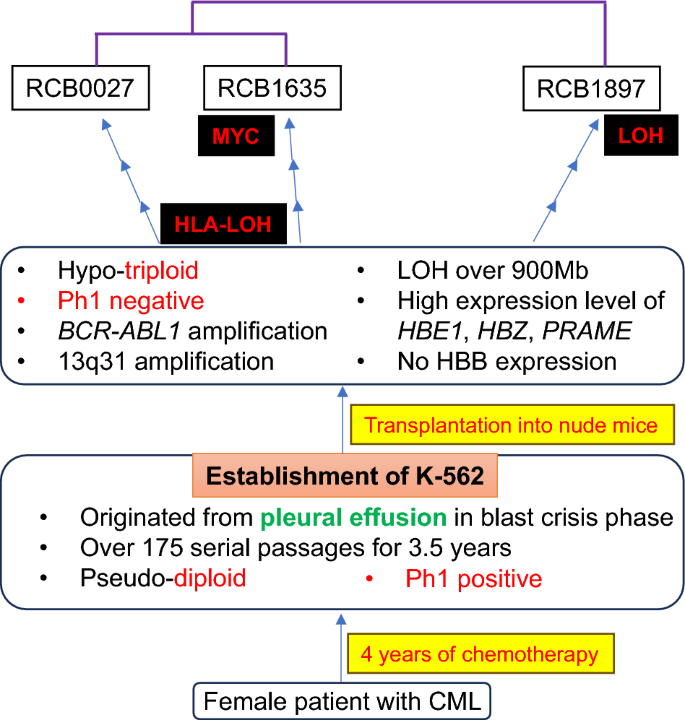
The study highlights genomic differences between three K562 sublines (RCB0027, RCB1635, and RCB1897), including variations in the BCR-ABL1 gene and loss of heterozygosity (LOH). These findings suggest that each subline has diverged significantly over time, influencing experimental outcomes in leukemia research.
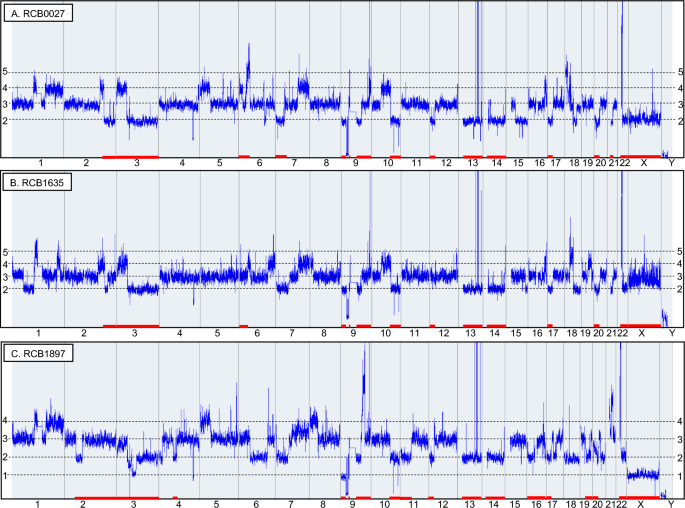 Figure 1: Copy number profiles compared among three K-562 sublines based on DNA microarray analysis.
Figure 1: Copy number profiles compared among three K-562 sublines based on DNA microarray analysis. -
Differentiation and Hemoglobin Synthesis:
Differences in hemoglobin synthesis and transcriptome profiles were observed between the K562 sublines. These variations emphasize the importance of selecting the right subline for studying leukemia’s molecular mechanisms and therapeutic responses. -
Implications for Leukemia Research:
This variability has critical implications for leukemia research, especially in the development of drug resistance models and gene editing applications. Researchers should recognize these sublines as distinct entities to avoid discrepancies in experimental data.
Figure 2 K-562 Fact Sheet, illustrating its fundamental cell data. The data consist of three parts: the clinical data, the establishment of the cell line, and current features.
Runtogen’s K562 Products for Advancing Leukemia Research
At Runtogen, we offer a variety of K562-based products designed to support cutting-edge research in CML, gene editing, and drug screening. Our offerings include:
-
K-562 Luciferase Reporter Cell Lines:
Ideal for bioluminescence assays, our luciferase-expressing K562 cells are highly sensitive and perfect for gene expression studies. -
K-562 Knockout Cell Lines:
Study gene function and disease modeling with our comprehensive range of K562 knockout cell lines. These models are crucial for investigating leukemia progression and therapy resistance. -
K-562 CRISPR-Cas9 Editing Tools:
Unlock the full potential of CRISPR gene editing in K562 cells. These models are perfect for studying targeted genetic changes and their impacts on leukemia biology. -
K-562 Drug Sensitivity Assays:
Our K562-based drug sensitivity assays help you discover novel anti-cancer compounds and evaluate drug resistance mechanisms in leukemia treatment.
Why Understanding K-562 Variability Matters in Leukemia Research
The study emphasizes that K562 cell line variability can lead to significant research discrepancies if sublines are not considered. Understanding the unique characteristics of each subline can enhance the reproducibility and reliability of leukemia research and improve therapeutic development.
By choosing the right K562 subline for your studies, you can ensure more accurate results and contribute to advancing leukemia therapy.
Explore Runtogen’s K562 Product Range
Explore our full collection of K562 cell lines and tools tailored for diverse applications in leukemia and cancer research. Visit our product page to learn more about how our K562 cell lines can help propel your research.
Explore K562 Products at Runtogen
Conclusion: The Role of K562 Cells in Modern Leukemia Research
The K562 cell line is more complex than previously assumed, with significant variability across sublines that can influence research outcomes. Researchers must carefully select the right subline to avoid discrepancies and improve research accuracy. At Runtogen, we offer a variety of tools to support your work, from gene editing to drug screening.
- K562 Cell Line Study: Scientific Reports (2024). “Variable characteristics overlooked in human K-562 leukemia cell lines with a common signature.” Nature Scientific Reports
- Zhou, B. et al. Comprehensive, integrated, and phased whole-genome analysis of the primary ENCODE cell line K562. Genome Res. 29, 472–484. https://doi.org/10.1101/gr.234948.118 (2019).